|
A wide extent of inter-strain diversity in virulent and vaccine strains of alpha-herpesviruses.
Szpara et al. PLoS Pathog 7(10): e1002282 |
| Home |
|---|
|
Abstract
|
| Explore |
|
A guide to this website, with links to related resources
|
| Scripts |
|
A guide to the scripts with instructions on their use
|
| Download |
|
Download links for data and files
|
| Supplement |
|
Supplementary Figures and Tables
|
| Authors |
|
|
| Download |
|---|
Table of Contents |
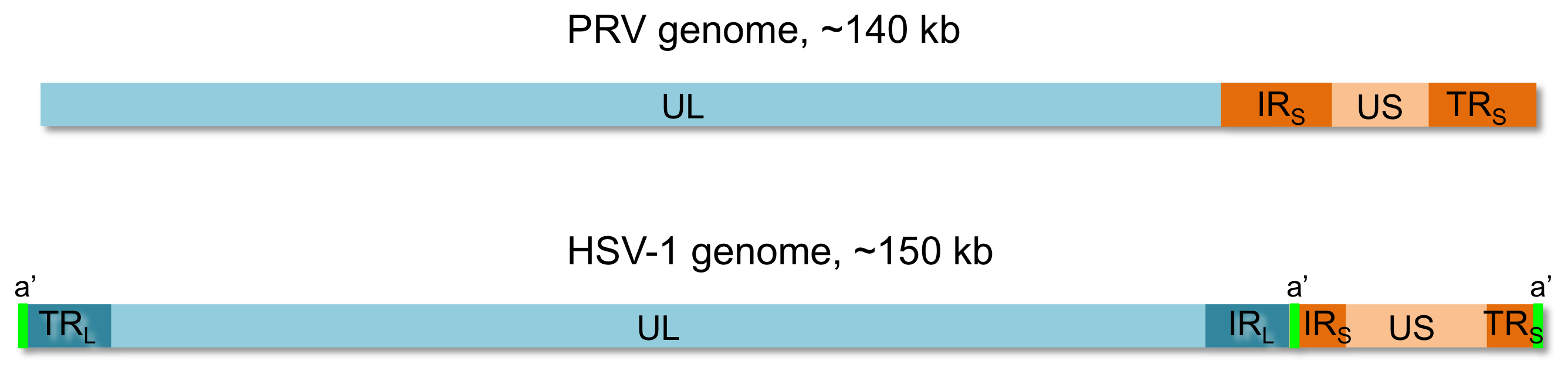
Assembly and Analysis ScriptsThe assembly and analysis scripts are available for download here, both individually and as a bundle. All scripts presented here were either written or adapted by Lance Parsons. If you download the scripts individually, you may want to make the Perl and Python scripts executable. Details about how the scripts were used are available in the the Scripts section of this website. The same information is in the downloadable file: Szpara_et_al_2011, Script Instructions (pdf) Gzipped bundle of scripts (.tar.gz):
|
Links to data deposited in online repositories |
Finished Genomes
|
SNP Data Files |
For questions about this website, send mail to the
Genomics Bioinformatics Group
at Princeton University.
Last updated: 27 December 2011