 |
Research Home
PNAS Article
PDF
Download Data
Related Work
- Coordination of Cell Growth and Division
- Growth Rate Response
- Decoupling of Cell Growth and Division
|
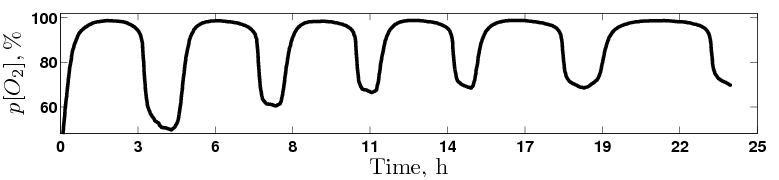
Despite rapid progress in characterizing the yeast metabolic cycle (YMC), its connection to the cell division cycle (CDC) has remained unclear.
We discovered that a prototrophic batch culture of budding yeast, growing in a phosphate-limited ethanol media, synchronizes spontaneously and goes through multiple metabolic
cycles while the fraction of cells in the G1/G0 phase of the CDC increases monotonically from 90% to 99%. This is the first example of metabolic synchrony without cell
division synchrony, and of YMC in non-carbon-source-limited cultures. More than 3000 genes, including most genes annotated to CDC, were expressed periodically in our batch culture
albeit mere 10% of the cells divided asynchronously; only a smaller subset of CDC genes correlated with cell division.
These results suggest that the YMC reflects a growth cycle during G1/G0 and allow extension and generalization of our mechanistic model
connecting gene expression in asynchronous cultures to the underlying YMC and CDC dynamics in single cells. The extended model explains our previous puzzling
observation that genes annotated to the CDC increase in expression at slow growth. We conclude that the life cycle of slowly growing yeast is comprised of distinct growth
and division cycles.
Phosphate-Limited Medium with Ethanol as a Sole Source of Carbon and Energy |
View Figures and Explore the Data Interactively |
All Genes Hierarchically Clustered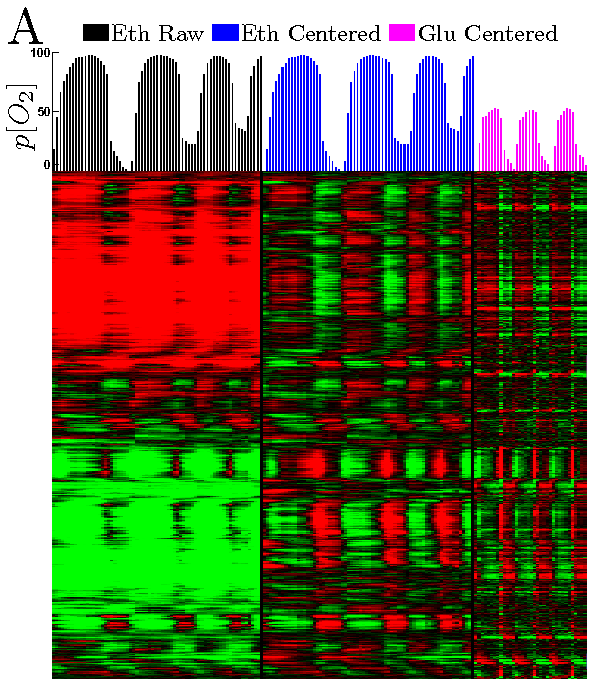
|
Phase-Ordered Genes Having the Same Phase in Both Batch Ethanol and in Continous Glucose Cultures
|
Genes Annotated to the Cell Division Cycle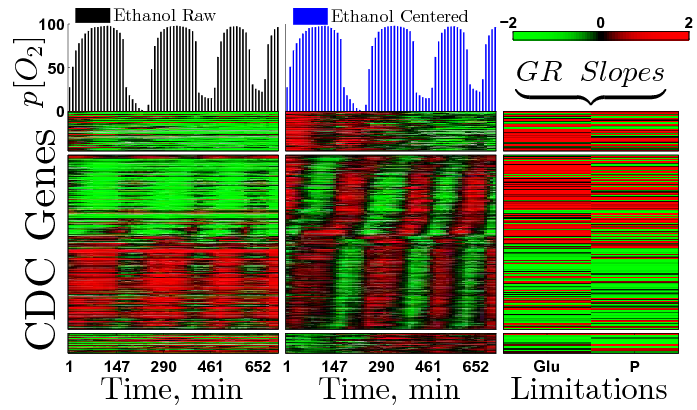
|